Home > Press > New class of protein misfolding simulated in high definition: Evidence for recently identified and long-lasting type of protein misfolding bolstered by atomic-scale simulations and new experiments
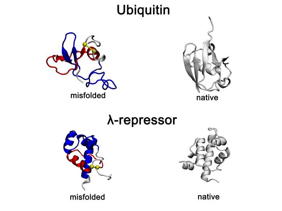 |
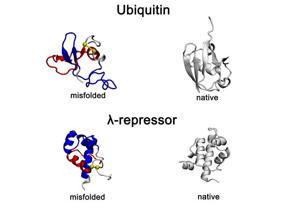
| Recently identified and long-lasting type of protein misfolding non-native entanglements observed in all-atom protein folding simulations. Representative misfolded conformations of the small proteins, Ubiquitin and λ-repressor, exhibit gains-of-entanglement in all-atom folding simulations and are shown alongside their native structures. In the misfolded states, non-native entangled loops are highlighted in red, with yellow spheres marking loop closures and blue segments indicating threading through the loop.
Credit OBrien Laboratory, Penn State |
Abstract:
New computer simulations that model every atom of a protein as it folds into its final three-dimensional form support the existence of a recently identified type of protein misfolding. Proteins must fold into precise three-dimensional shapes called their native state to carry out their biological functions. When proteins misfold, they can lose function and, in some cases, contribute to disease. The newly spotted misfolding results in a change to a proteins structure either a loop that traps another section of the protein forms when it shouldnt or doesnt when it should that disrupts its function and can persist in cells by evading the cells quality control system. The simulated misfolds also align closely with structural changes inferred from experiments that track protein folding using mass spectrometry, according to the team led by researchers at Penn State.
New class of protein misfolding simulated in high definition: Evidence for recently identified and long-lasting type of protein misfolding bolstered by atomic-scale simulations and new experiments
University Park, PA | Posted on August 8th, 2025
Protein misfolding can cause disease, including Alzheimers and Parkinsons, and is thought to be one of the many factors that influence aging, said Ed OBrien, professor of chemistry in the Eberly College of Science, a co-hire of the Institute for Computational and Data Sciences at Penn State and the leader of the research team. This research represents another step forward in our attempt to document and understand the mechanisms of protein misfolding. Our aim is to translate these fundamental discoveries into therapeutic targets that could help mitigate the impacts of these disorders and even aging.
A paper describing the research appeared today (Aug. 8) in the journal Science Advances.
Proteins are composed of long strings of units called amino acids. A proteins function relies on the sequence of those amino acids along the string, which determines how the string will fold into a three-dimensional structure. Sections of the protein can fold into helices, loops, sheets and various other structures which allows them to interact with other molecules and perform their functions. Any mistake during this folding process can disrupt these functions.
The new class of misfolding, recently identified by the OBrien Lab, involves a change in entanglement status in the proteins structure. Entanglement refers to sections of the string of amino acids looping around each other like a lasso or a knot. Sometimes an entanglement can form when it shouldnt be there and sometimes an entanglement that is part of the proteins native structure doesnt form when it should.
In our previous study, we used a coarser-grained simulation that only modeled the protein at the amino acid level not the atomic level, said Quyen Vu, first author of the paper and a postdoctoral researcher in chemistry at Penn State who started the research as a graduate student at the Polish Academy of Sciences. But there was concern in the community that such a model might not be realistic enough, as the chemical properties and bonding of the atoms that make up amino acids influence the folding process. So, we wanted to make sure we still saw this class of entanglement misfolding with higher-resolution simulations.
The team first used all-atom models of two small proteins and simulated their folding. They found that both small proteins could form the misfolds just like in their coarser-grained simulations. However, unlike in their previous simulations, which modeled normal-sized proteins, the misfolds in these small proteins lasted only a short time.
We think that the misfolds in our previous simulations persisted for two main reasons, Vu said. First, to fix the misfold required backtracking and unfolding several steps to correct to entanglement status, and second, the misfold can be buried deep inside the proteins structure and essentially invisible to the cells quality control system. With the small proteins there were fewer steps and less to hide behind so the mistakes could be quickly fixed. So, we simulated a normal size protein at the atomic scale and saw misfolding that persisted.
The team also tracked folding of the proteins used in their simulations experimentally. While they couldnt directly observe the misfolds in the experiments, structural changes inferred using mass spectrometry occurred in the locations that misfolded in their simulations.
Most misfolded proteins are quickly fixed or degraded in cells, OBrien said. But this type of entanglement presents two major problems. They are difficult to fix as they can be very stable, and they can fly under the radar of the cells quality control systems. Coarse-grain simulations suggest that this type of misfolding is common. Learning more about the mechanism can help us understand its role in aging and disease and hopefully point to new therapeutic targets for drug development.
In addition to Vu and OBrien, the research team includes Ian Sitarik, graduate student in chemistry; Yang Jiang, assistant research professor in chemistry; and Hyebin Song, assistant professor of statistics, at Penn State; Yingzi Xia, Piyoosh Sharma, Divya Yadav, and Stephen D. Fried at Johns Hopkins University; and Mai Suan Li at the Polish Academy of Sciences.
The U.S. National Science Foundation, the U.S. National Institutes of Health and the Polish National Science Centre funded the research. The research was supported in part by the TASK Supercomputer Center in Gdansk, Poland; the PLGrid Infrastructure in Poland; and the Roar supercomputer in the Institute for Computational and Data Sciences at Penn State.
####
For more information, please click here
Contacts:
Media Contact
Sam Sholtis
Penn State
Office: 814-865-1390
Expert Contact
Edward O’Brien
Penn State
Copyright © Penn State
If you have a comment, please Contact us.
Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
News and information
![]()
Sensors innovations for smart lithium-based batteries: advancements, opportunities, and potential challenges August 8th, 2025
![]()
Deciphering local microstrain-induced optimization of asymmetric Fe single atomic sites for efficient oxygen reduction August 8th, 2025
![]()
Lab to industry: InSe wafer-scale breakthrough for future electronics August 8th, 2025
![]()
New imaging approach transforms study of bacterial biofilms August 8th, 2025
Possible Futures
![]()
ICFO researchers overcome long-standing bottleneck in single photon detection with twisted 2D materials August 8th, 2025
![]()
New molecular technology targets tumors and simultaneously silences two undruggable cancer genes August 8th, 2025
![]()
Simple algorithm paired with standard imaging tool could predict failure in lithium metal batteries August 8th, 2025
![]()
First real-time observation of two-dimensional melting process: Researchers at Mainz University unveil new insights into magnetic vortex structures August 8th, 2025
Discoveries
![]()
Deciphering local microstrain-induced optimization of asymmetric Fe single atomic sites for efficient oxygen reduction August 8th, 2025
![]()
ICFO researchers overcome long-standing bottleneck in single photon detection with twisted 2D materials August 8th, 2025
![]()
New molecular technology targets tumors and simultaneously silences two undruggable cancer genes August 8th, 2025
![]()
Simple algorithm paired with standard imaging tool could predict failure in lithium metal batteries August 8th, 2025
Announcements
![]()
Sensors innovations for smart lithium-based batteries: advancements, opportunities, and potential challenges August 8th, 2025
![]()
Deciphering local microstrain-induced optimization of asymmetric Fe single atomic sites for efficient oxygen reduction August 8th, 2025
![]()
Japan launches fully domestically produced quantum computer: Expo visitors to experience quantum computing firsthand August 8th, 2025
![]()
ICFO researchers overcome long-standing bottleneck in single photon detection with twisted 2D materials August 8th, 2025
Interviews/Book Reviews/Essays/Reports/Podcasts/Journals/White papers/Posters
![]()
New molecular technology targets tumors and simultaneously silences two undruggable cancer genes August 8th, 2025
![]()
Simple algorithm paired with standard imaging tool could predict failure in lithium metal batteries August 8th, 2025
![]()
First real-time observation of two-dimensional melting process: Researchers at Mainz University unveil new insights into magnetic vortex structures August 8th, 2025
![]()
Lab to industry: InSe wafer-scale breakthrough for future electronics August 8th, 2025